현재 위치:홈 > 뉴스현황 > Press Events > Virtual Embryo Predi...
저자: 업로드:2017-09-06 조회수:
By the time an embryo starts to show form and structure, every cell in the embryo “knows” its fate, whether it is to become a brain cell, a heart cell, or any other kind of cell. It already has, at this early stage of development, a unique transcriptional identity, one that reflects the cell’s position within the embryonic whole, one that follows a transcriptional blueprint.
Now scientists are finally getting a peek at transcriptional blueprints, starting with the spatial gene expression patterns for Drosophila melanogaster. The scientists, based at the Max Delbrück Center (MDC), have analyzed the unique gene expression profiles of thousands of single cells and reassembled the embryo from these data using a new spatial mapping algorithm. The result is a virtual fly embryo showing exactly which genes are active where at a crucial stage in morphogenesis, stage 6, the onset of gastrulation.
The D. melanogaster embryo, the MDC scientists explained, has been an exquisite model for the patterning principles that shape cellular identities: “The fertilized egg undergoes 13 rapid nuclear divisions resulting in a syncytial embryo of about 6000 nuclei. By developmental stage 5, nuclei have moved to the embryo periphery, become surrounded by cell membranes, and spatial gene expression patterns emerge as cells translate anteroposterior and dorsoventral positional information into transcriptional responses. Stage 6 is marked by the first morphogenetic movements after cellularization completes.”
To capture patterning principles, the MDC scientists devised a computational mapping strategy called DistMap. Using this strategy, the scientists reconstruct the embryo to predict spatial gene expression approaching single-cell resolution. Details of this work appeared August 31 in the journal Science, in an article entitled “The Drosophila Embryo at Single-Cell Transcriptome Resolution.”
“We produce a virtual embryo with about 8000 expressed genes per cell,” the article’s authors wrote. “Our interactive “Drosophila-Virtual-Expression-eXplorer” (DVEX) database generates three-dimensional virtual in situ hybridizations and computes gene expression gradients.”
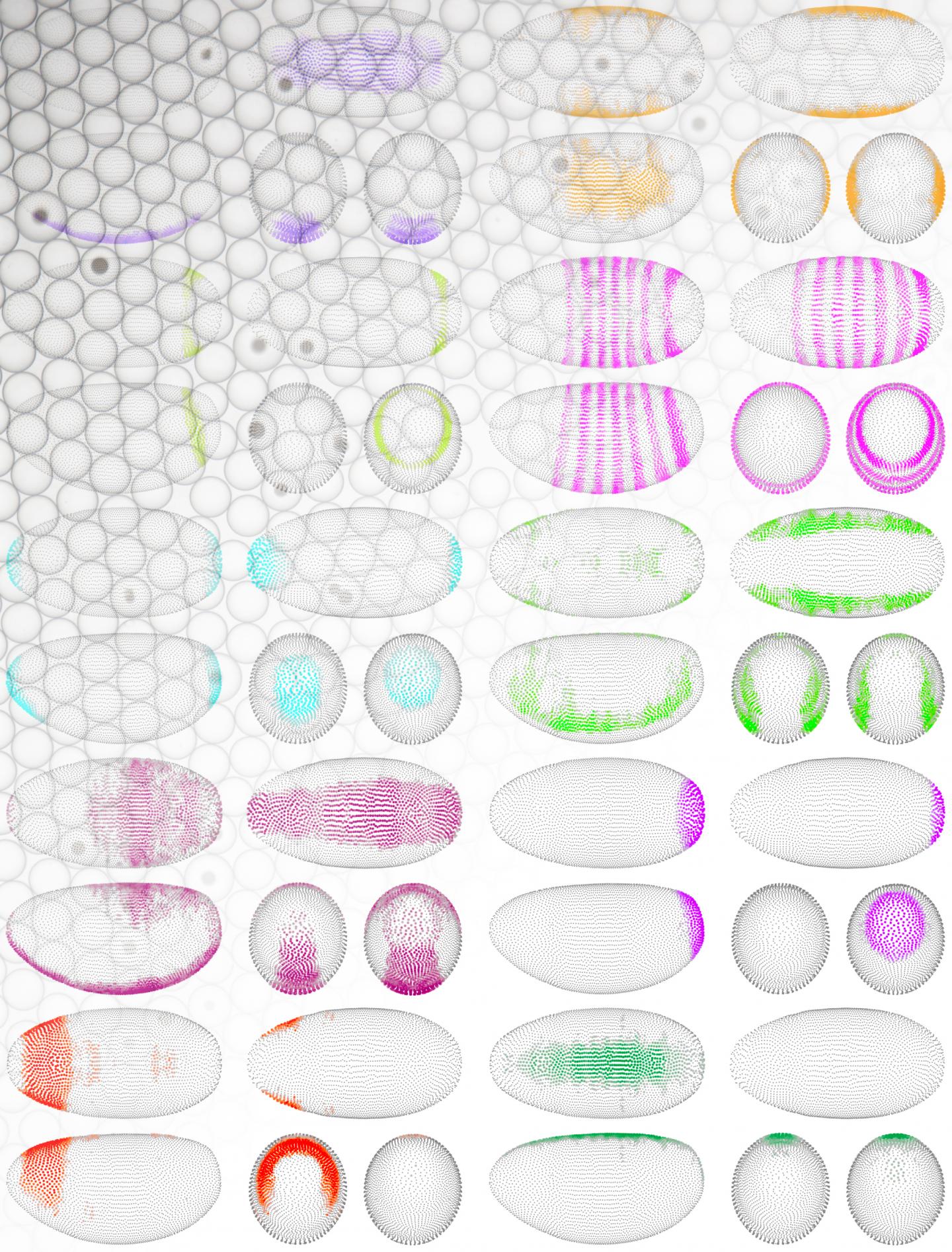
Essentially, the MDC team computed genome-wide spatial gene expression patterns from single-cell transcriptome data alone. In their paper, the MDC researchers describe a dozen new transcription factors and many more long noncoding RNAs that have never been studied before. Also, they propose an answer to a question that has puzzled scientists for 35 years: How does the embryo break synchronicity of cell divisions to develop more comple