현재 위치:홈 > 뉴스현황 > Press Events > Gene Synthesis Upgra...
저자: 업로드:2017-07-19 조회수:
Protein designers want to venture boldly into uncharted protein space, but they tend to explore only so far as their computational models and experimental screens allow. Essentially, protein designers hug the shore of what is known about the folding of native proteins. Unfortunately, this approach can take protein designers in the wrong direction. Native proteins have evolved for function, not stability, so “natural” protein space may give a limited view of the connections between amino acid sequences and folded, three-dimensional protein structures.
A broader view, one that would survey natural and unnatural protein structures, would be instructive, but to date, screens of computationally derived structures have been limited to 50 to 100 designs. Screens of thousands, however, have been demonstrated by scientists based at the University of Toronto and the University of Washington. These scientists, led by Toronto’s Cheryl Arrowsmith, Ph.D., and Washington’s David Baker, Ph.D., have taken data-driven, high-throughput protein design well beyond the old protein-folding coastline.
The researchers tested more than 15,000 newly designed miniproteins that do not exist in nature to see whether they form folded structures. "We learned a huge amount at this new scale, but the taste has given us an even larger appetite," said Gabriel Rocklin, Ph.D., a researcher in Dr. Baker’s laboratory. "We're eager to test hundreds of thousands of designs in the next few years."
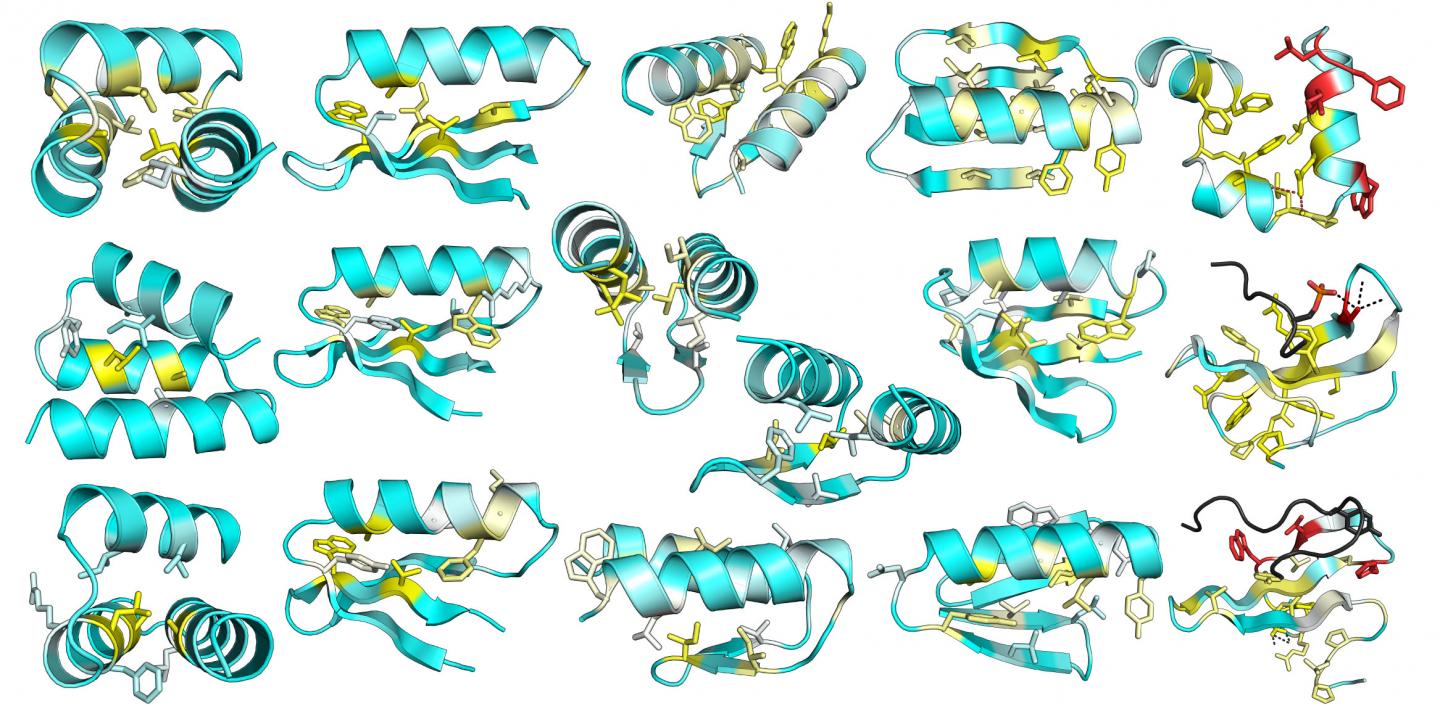
Details of this work appeared July 14 in the journal Science, in an article entitled “Global Analysis of Protein Folding Using Massively Parallel Design, Synthesis, and Testing.” This article describes a testing project that led to the design of 2788 stable protein structures and could have many bioengineering and synthetic biology applications.
“We combined computational protein design, next-generation gene synthesis, and a high-throughput protease susceptibility assay to measure folding and stability for more than 15,000 de novo designed miniproteins, 1000 natural proteins, 10,000 point mutants, and 30,000 negative control sequences,” wrote the article’s authors. “This analysis identified more than 2500 stable designed proteins in four basic folds—a number sufficient to enable us to systematically examine how sequence determines folding and stability in uncharted protein space.”
For decades, researchers have studied protein folding by examining the structures of naturally occurring proteins. However, natural protein structures are typically large and complex, with thousands of interactions that collectively hold the protein in its folded shape. Measuring the contribution of each interaction becomes very difficult.